Investigation of Protein-Protein Interactions with the help of the Strep-tag® technology
Understanding protein-protein interactions (PPIs) is fundamental to the field of molecular biology. These interactions regulate crucial biological processes, from signal transduction and immune responses to maintaining cellular structure and function. Many diseases are linked to protein interactions. By investigating these interactions, researchers can identify potential therapeutic targets, understand disease mechanisms, optimize leads during the drug discovery process, or discover biomarkers for disease diagnosis and monitoring.
The key steps to identify and characterize the interaction between two specific proteins include:
- Experimental Design: Define the target protein and choose the appropriate methods based on the specific goals and available resources
- Lead Identification: Collect data to identify potential interaction partners. This step involves screening and identifying promising leads that show significant interaction.
- Confirmation: Validate initial findings using complementary techniques in-vitro or in-vivo.
- Characterization: Further characterize the interaction structurally and functionally, for example by measuring binding kinetics in SPR
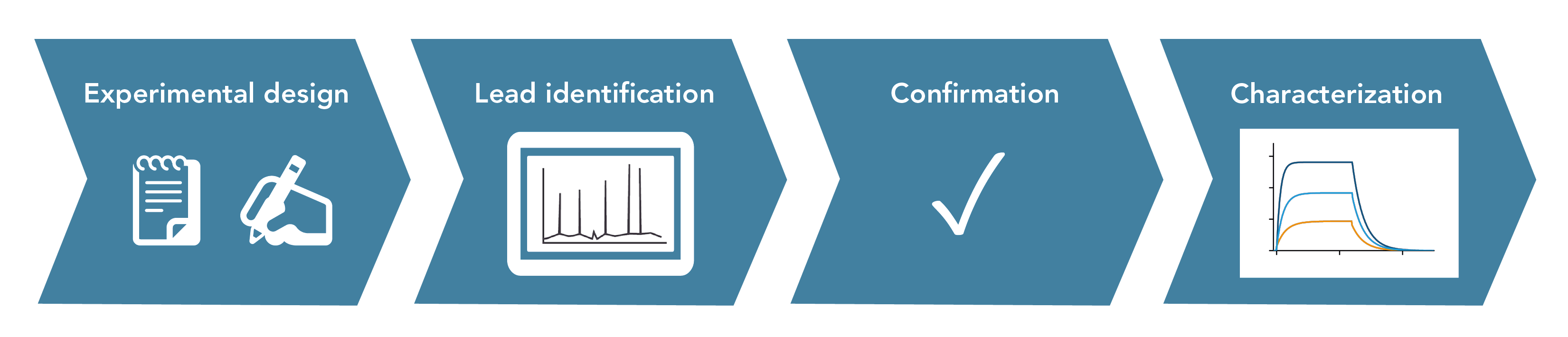
Figure 1: Typical workflow of an experiment to identify and analyze protein-protein interactions
Experimental Design: Selecting experimental conditions
To ensure an easy workflow, as well as accurate and reliable results, several factors should be considered while planning the experimental setup. Selecting the appropriate detection method, protein expression system and conditions for sample preparation are vital. Incorporating protein tags, such as Twin-Strep-tag®, His-tag or FLAG-tag can offer advantages.
Protein tags facilitate the purification and detection of target proteins and can improve the efficiency of the experiment. They can also aid in further steps, like the ability to immobilize the protein during analysis steps, for example in SPR. To offer benefits to the analysis workflow, the ideal tag should have the following properties:
- No unspecific interaction
- High affinity
- Little to no influence on protein structure
- Small size
- Reversible binding
- Multiple possible applications
Among the most commonly used tags, the Twin-Strep-tag® possesses all these properties and offers the greatest benefit for PPI analysis.
Lead identification: Affinity chromatography, mass spectrometry and bioinformatics
After a target protein has been chosen, the possible interaction partners have to be identified. Isolating the target protein from the cell while keeping its interactions intact is a convenient way to do this. Pull-down assays and Co-immunoprecipitation (Co-IP) are two effective methods for detecting protein-protein interactions. Co-IP utilizes immobilized antibodies to purify the bait protein along with the prey from cell lysates, enabling the analysis of the entire protein complex. In contrast, pull-down assays involve fusing the target protein to an affinity tag and capturing it, along with any bound proteins, using a corresponding immobilized ligand.
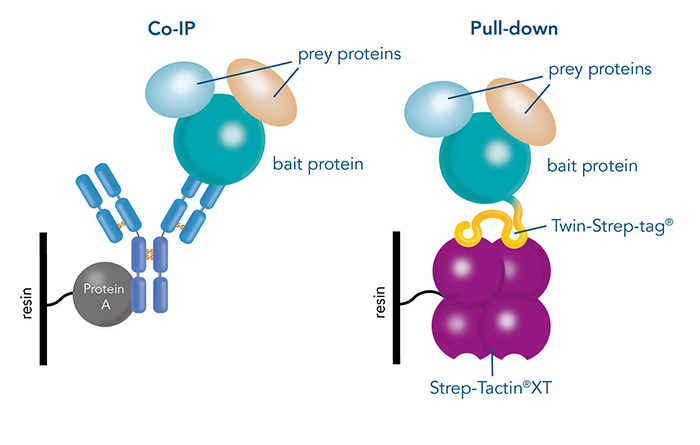
Figure 2: Differences between Co-IP via protein specific antibodies and a pull-down, which is performed via a specific protein tag.
While antibodies are a great choice when specific ones antibodies are available, this is not always the case. In addition, specificity and binding affinity can vary between different antibodies, leading to unreliable results. Pull-down assays, for example via Twin-Strep-tag® and Strep-Tactin®XT offer several advantages. The same tag and ligand system can be used for different proteins, while the same affinity and specificity is guaranteed. Furthermore, they offer mild elution conditions, which preserves the interaction between the bait and prey proteins.
In a subsequent step, the potential interaction partners are then identified, for example by mass spectrometry. These experimental methods are complemented by bioinformatic methods, predicting potential interaction partners based on sequence and structural data from databases. Combining computational and experimental procedures enhances the accuracy and efficiency of lead identification, allowing researchers to narrow down the pool of candidates that need to be screened. Once the most promising leads are identified, each interaction must be individually studied for detailed characterization to understand the biological significance and mechanism of the interactions.
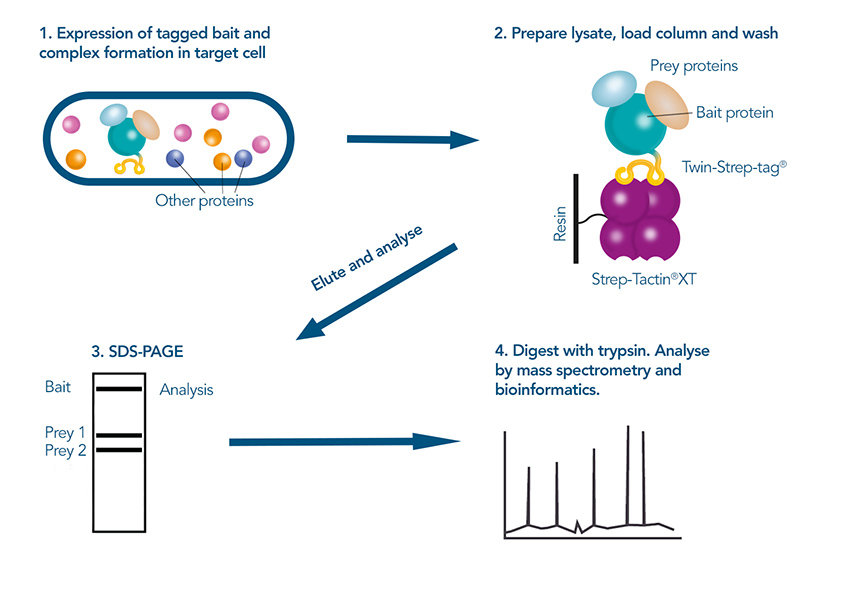
Figure 3: Using a pull-down-assay to identify protein interactions: The tagged target protein is expressed in the host cell, after which it is purified under mild conditions via the Twin-Strep-tag®. Prey proteins that were co purified are separated from the bait protein in gel electrophoresis, digested and then identified in mass spectrometry.
Confirming the interaction
After potential interaction partners have been identified, further steps are required to confirm the interaction. For this, the original bait protein and the prey proteins are recombinantly produced with modifications that facilitate the detection of their interaction.
One approach involves labeling the proteins with agents that generate a signal when brought into close proximity by the interaction. These agents can be parts of proteins or specific fluorophores. For example, in Homogeneous Time-Resolved Fluorescence (HTRF) assays, donor and acceptor fluorophores are conjugated to molecules that recognize the binding partners, such as Strep-Tactin® or antibodies. This assay relies on Förster resonance energy transfer (FRET), where energy is transferred from a donor fluorophore to an acceptor fluorophore and induces a signal when they are close together. HTRF offers high sensitivity and reduced background noise through time-resolved measurement
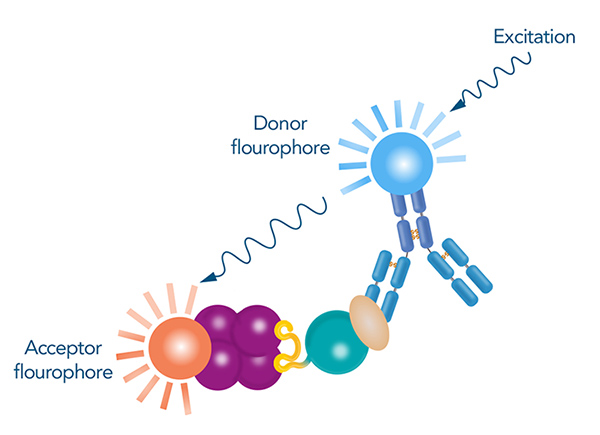
Figure 4: Principle of HTRF for confirmation of protein interactions. The donor fluorophore is conjugated to an antibody specific to the prey protein, while the acceptor fluorophore is conjugated to a ligand that binds to the tag of the bait protein. The donor fluorophore is excited by a light of a certain wavelength and transfers its energy to the acceptor fluorophore if they are in close proximity, which results in a measurable signal.
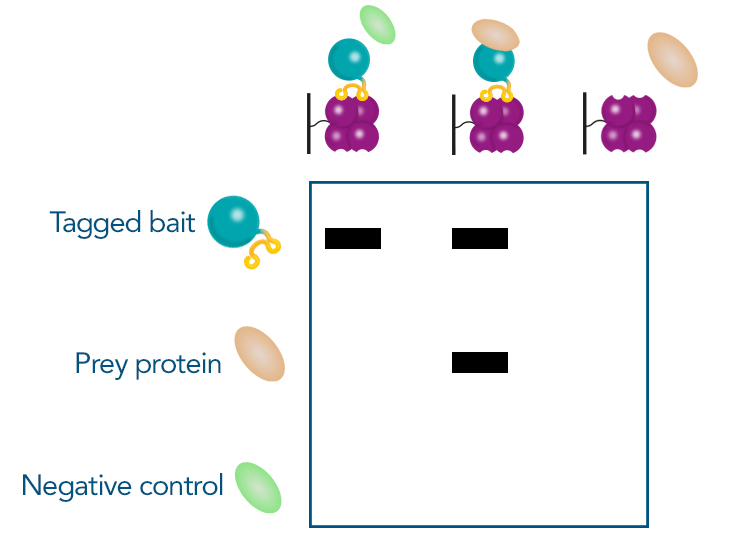
Another straightforward method, especially if the initial screening was performed via a pull-down assay, is a subsequent pull-down. In this method, the chosen prey protein is recombinantly expressed and mixed with the bait protein. An affinity purification targeting the bait protein is then conducted. The presence of the prey protein is confirmed by Western blot analysis, comparing results to negative controls to validate the interaction. Other methods not only confirm the interaction but also provide additional insights. For example, protein crystallography can determine protein structure, or Surface Plasmon Resonance (SPR) can measure binding kinetics. These advanced techniques offer detailed information on the nature and dynamics of the interaction.
Figure 5: Confirmation of protein interaction in SDS page after Pull-down-assay. Prior to the experiment, the tagged bait protein, prey protein and an unrelated control protein are recombinantly expressed. The proteins are then mixed in different combinations and purified via the tag: The bait protein with the control protein, the bait protein with the prey protein and the prey protein on its own. If the prey protein is only visible in the gel if it was mixed with the bait protein, the interaction is confirmed.
Characterizing the interaction: SPR and BLI
Once an interaction partner has been identified and confirmed, the interaction can be further characterized to gain deeper insights into its biological function. Factors such as binding kinetics and affinity are crucial for understanding the role of the interaction in the organism. Techniques like surface plasmon resonance (SPR) and biolayer interferometry (BLI) are particularly useful for this purpose.
Both SPR and BLI offer real-time analysis of protein interactions, allowing for precise measurement of association and dissociation rates. Both techniques require the immobilization of the bait protein on a sensor or chip surface in a stable and oriented manner. The Twin-Strep-tag® technology is highly advantageous for this step due to its picomolar affinity, ensuring secure and directional immobilization of the bait protein. This allows the measurement of interactions of low to high affinites. Moreover, surfaces coated with Strep-Tactin®XT can be regenerated and reused multiple times, making these biosensor platforms cost-effective.
By utilizing the strengths of SPR and BLI, along with the robust immobilization provided by affinity tags like the Twin-Strep-tag®, researchers can obtain detailed kinetic and affinity data. This information is essential for revealing the functional roles of protein-protein interactions and their potential implications in cellular processes and disease mechanisms.
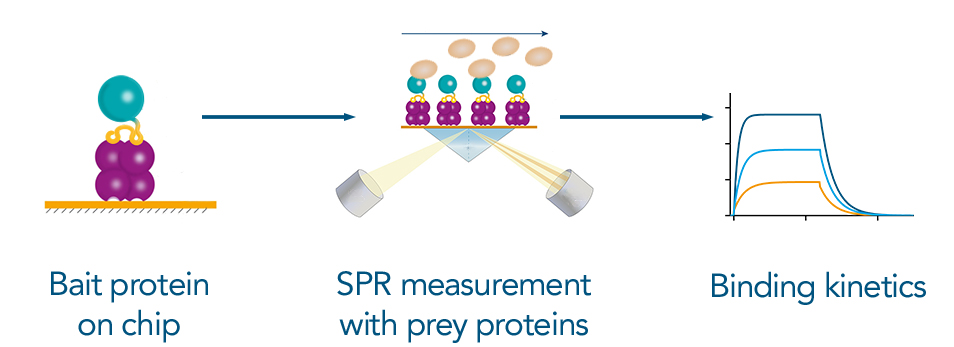
Figure 6: Measuring Binding kinetics via SPR: The tagged bait protein can bind to the SPR chip via the high affinity Twin-Strep-tag® to Strep-Tactin®XT. The prey protein is applied to the chip for the measurement. The resulting binding kintetics are essential information to characterize an interaction between two proteins.
References
- Gordon, D.E., Jang, G.M., Bouhaddou, M. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020). https://doi.org/10.1038/s41586-020-2286-9
- He L, Valignat MP, Zhang L, Gelard L, Zhang F, Le Guen V, Audebert S, Camoin L, Fossum E, Bogen B, Wang H, Henri S, Roncagalli R, Theodoly O, Liang Y, Malissen M, Malissen B. ARHGAP45 controls naïve T- and B-cell entry into lymph nodes and T-cell progenitor thymus seeding. EMBO Rep. 2021 Apr 7;22(4):e52196. https://doi.org/10.15252/embr.202052196
- Hou YN, Cai Y, Li WH, He WM, Zhao ZY, Zhu WJ, Wang Q, Mai X, Liu J, Lee HC, Stjepanovic G, Zhang H, Zhao YJ. A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1. Nat Commun. 2022 Dec 22;13(1):7898.
https://doi.org/10.1038/s41467-022-35581-y