Cloning
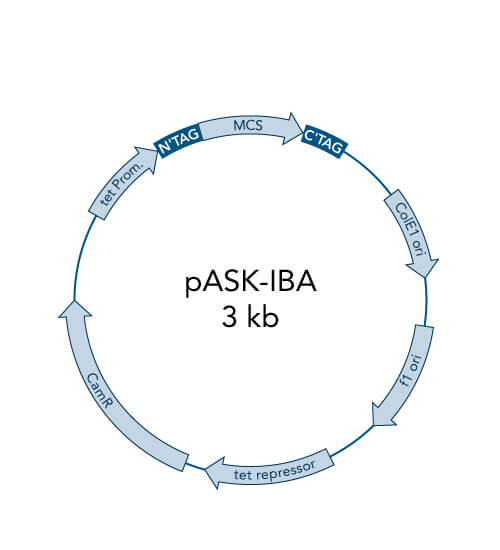
The objective of molecular cloning procedures is to introduce a gene of interest into a suitable expression host in a compatible manner (recombinant DNA) to ultimately produce the gene product (protein) in satisfying quality and quantity. Cloning means producing identical copies of a certain gene of interest. By cloning, the gene is amplified and then inserted into a plasmid (vector) for the following replication and protein expression. Using plasmids allows for the protection of the foreign genetic material from the expression host’s innate degradation machinery.
There are different cloning procedures used to insert the gene of interest into a plasmid. Restriction enzyme-based cloning is the standard cloning method in molecular biology. Restriction endonucleases cleave double-stranded DNA (dsDNA) at specific sequence sites called recognition sites. Depending on the employed restriction enzyme, the generated DNA fragments can either have blunt ends or sticky ends. These can be fused with plasmid DNA if it is cleaved with the same restriction enzyme and thus linearized. The fusion of the DNA fragment and the linearized plasmid DNA takes place with the help of a so-called DNA ligase.
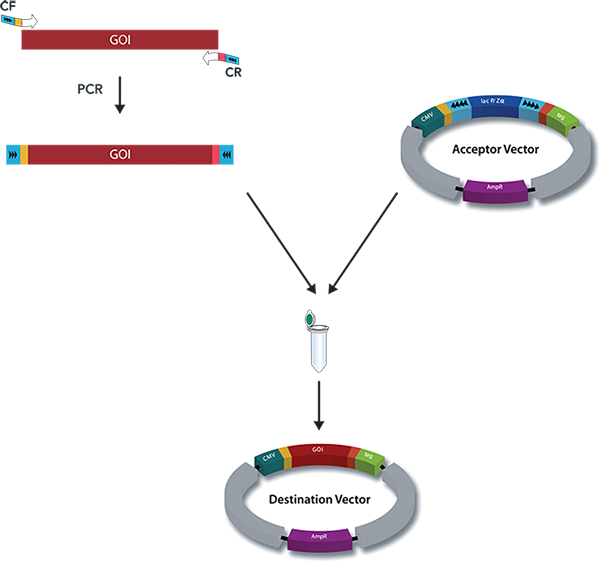
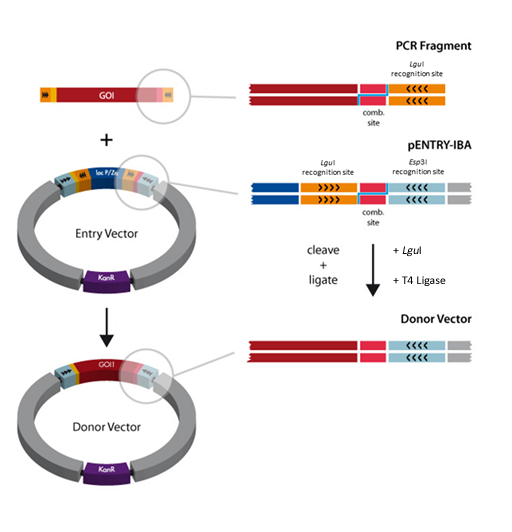
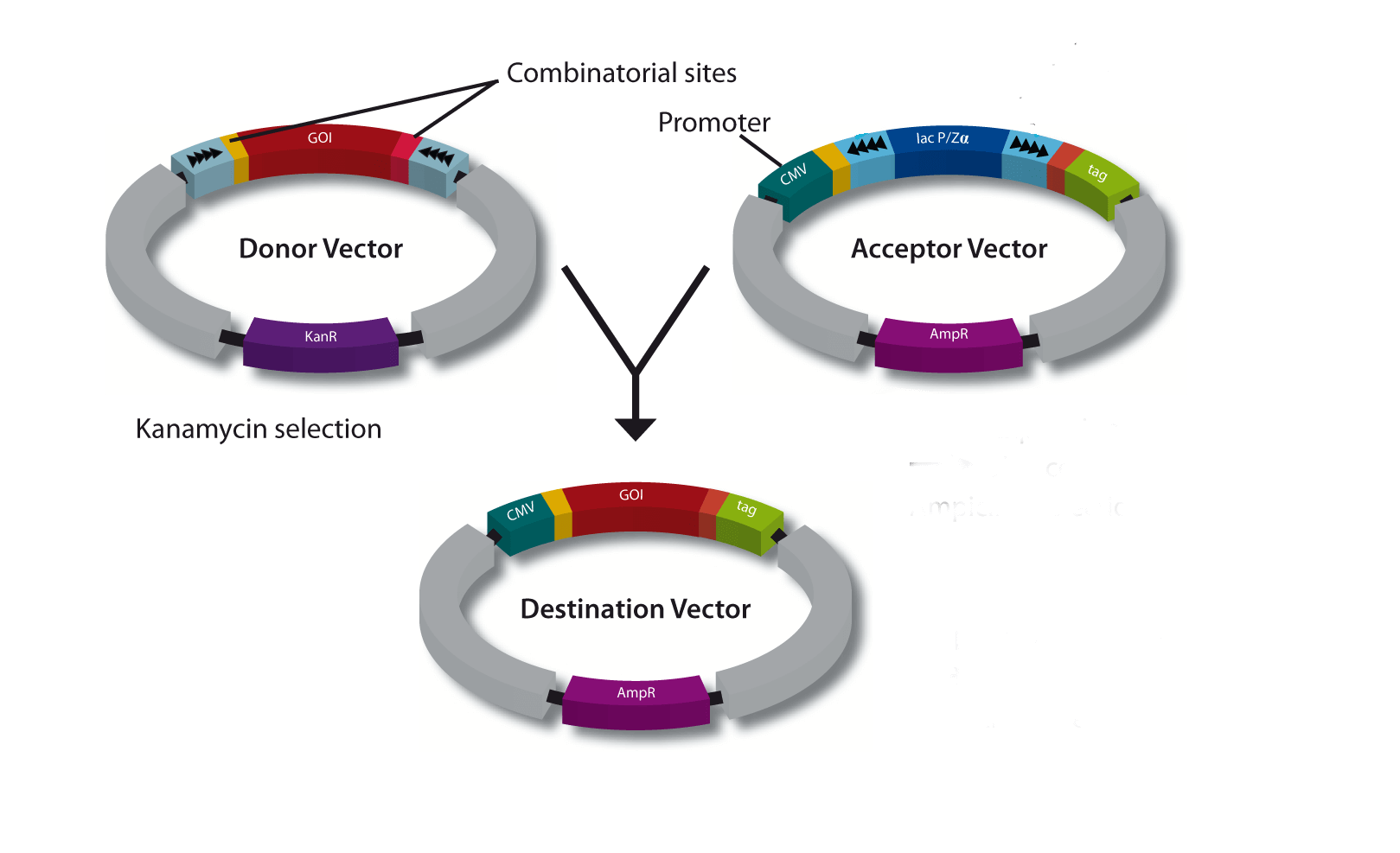
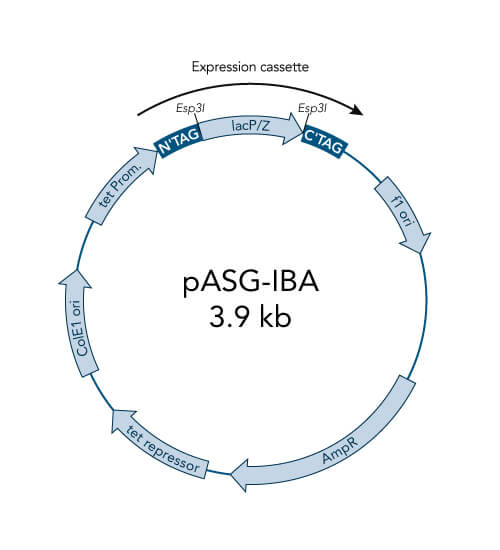
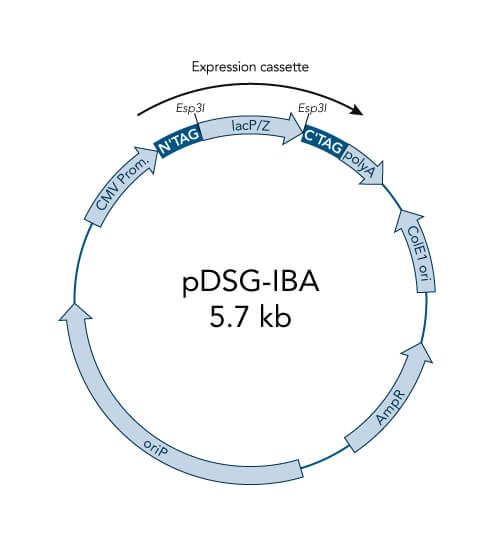
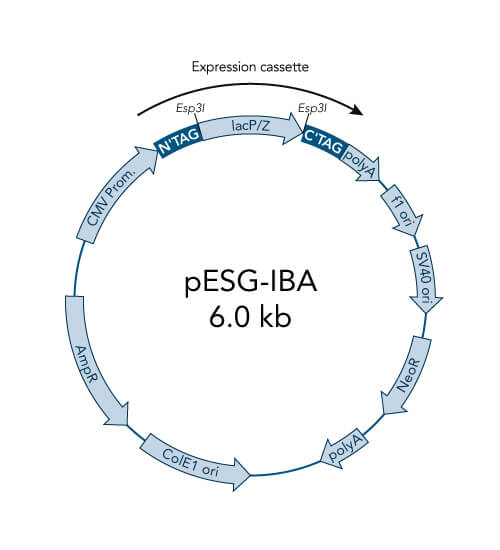
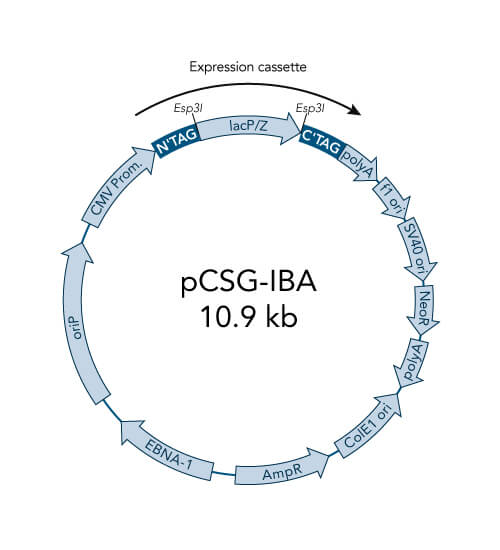
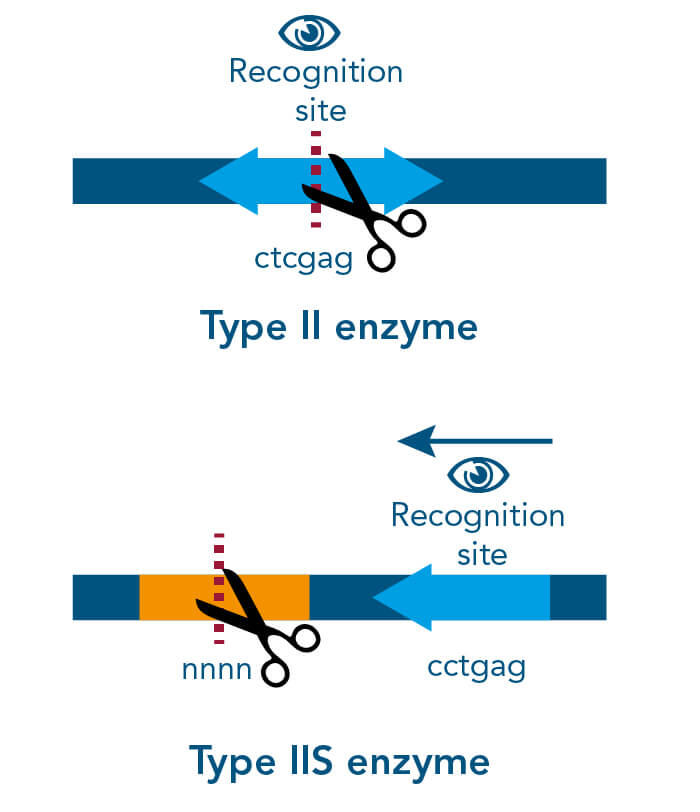
A special restriction enzyme-based cloning method makes use of the specific properties of type IIS restriction enzymes, endonucleases that cleave DNA outside of the recognition sequence. In contrast to traditional restriction enzyme cloning, this cutting outside from the recognition site provides the advantage that custom overhangs can be generated. With the StarGate system, IBA has utilized this approach to create compatible custom overhangs, which can then be fused efficiently. The StarGate system comes along with a large subset of cloning and expression vectors for E. coli or mammalian cells.
StarGate for fast protein expression cloning
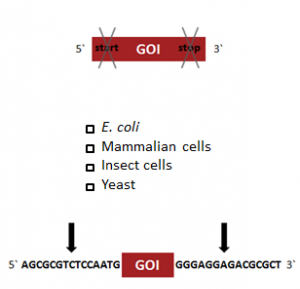
StarGate has been developed for the rapid systematic screen of the optimal expression (regarding expression host and/or promoter) and purification (regarding the fusion tag) system for a given gene of interest (GOI).
There are two possible cloning procedures.